We employ various visual and interactive tools for the analysis. Following are some of the selected fields in which we provide analysis:
We employ various visual and interactive tools for the analysis. Following are some of the selected fields in which we provide analysis:




HT-SELEX (High Throughput Systematic Evolution of Ligands by EXponential enrichment), SELEX-seq (Systematic Evolution of Ligands by EXponential enrichment with massively parallel sequencing), MITOMI (Mechanically Induced Trapping Of Molecular Interactions), HiTS-FLIP (High-Throughput Sequencing Fluorescent Ligand Interaction Profiling), Bind-n-Seq, CSI (Cognate Site Identification) array , CSI-Seq (Cognate Site Identification by Sequencing) & PBM (Protein Binding Microarray) are few of the methods to measure DNA protein interaction in vitro. SEQRS is a method that integrates in vitro selection, high-throughput sequencing of RNA, and SSLs (sequence specificity landscapes) for measurement of RNA binding of protein in vitro.
Artificial Transcription Factors (ATFs) like - polyamides, TALEs, Zinc-Fingers, etc. are created artificially to target a particular DNA and RNA sequence. COSMIC (Crosslinking Of Small Molecules for Isolation of Chromatin) and COSMIC by Sequencing (COSMIC-Seq) are some of the methods developed by us to measure in vivo binding of polyamides.
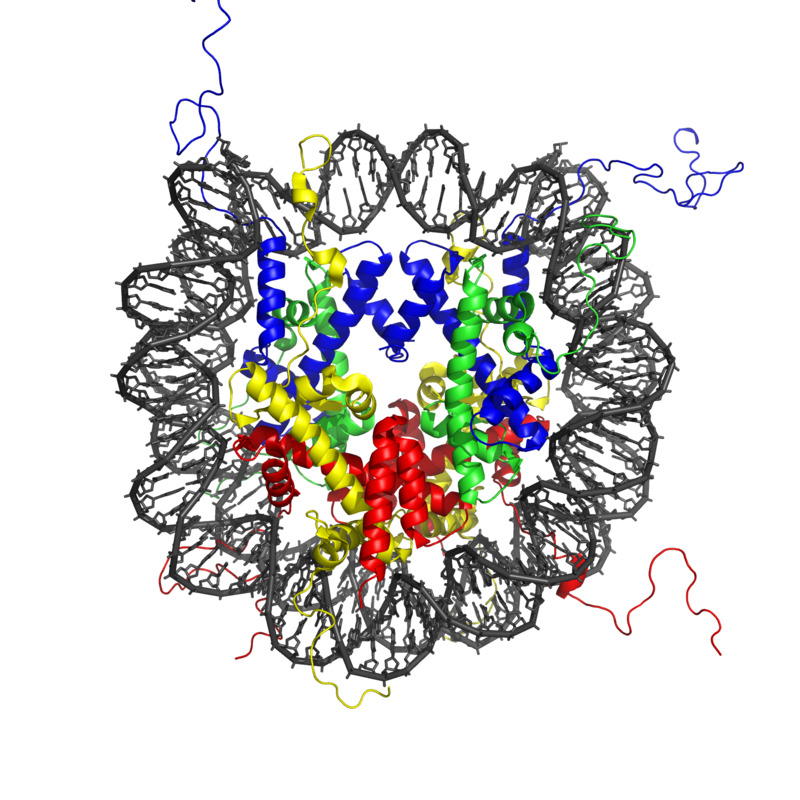
Chromatin immunoprecipitation followed by sequencing (ChIP-seq) is an experimental method used to measure in vivo DNA protein interactions.
Single Nucleotide Polymorphisms (SNPs) in non-coding region of genome can cause diseases by disrupting DNA binding TFs.
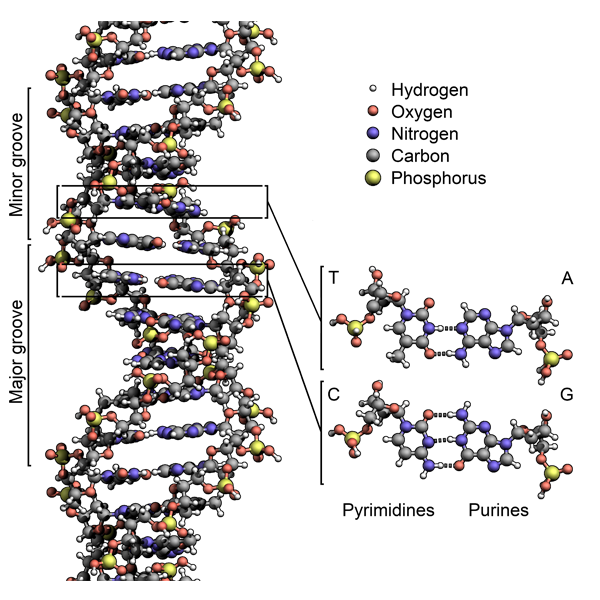
DNA binding of many TFs and proteins get affected by the shape and structure corresponding to different DNA sequences.
We develop novel methods to represent and analyze DNA and RNA binding motifs and patterns like effect of sequences flanking the main motif and binding to sub-maximal sites.
We build software packages, programs and tools for various bioinformatics analysis.
Specificity and Energy Landscapes (SELs) and other interactive tools and packages are developed by us to visualize the HT DNA/RNA binding data.
We analyze and create various models to integrate and explain relation between various HT datasets like ChIP, COSMIC, RNAseq, SNPs, etc. For example, sum of sites (SOS) model was created to integrate COSMIC and CSI-Seq.